Set up a network containing individual patient data (IPD). Multiple data
sources may be combined once created using combine_network().
Usage
set_ipd(
data,
study,
trt,
y = NULL,
r = NULL,
E = NULL,
Surv = NULL,
trt_ref = NULL,
trt_class = NULL
)Arguments
- data
a data frame
- study
column of
dataspecifying the studies, coded using integers, strings, or factors- trt
column of
dataspecifying treatments, coded using integers, strings, or factors- y
column of
dataspecifying a continuous outcome- r
column of
dataspecifying a binary outcome or Poisson outcome count- E
column of
dataspecifying the total time at risk for Poisson outcomes- Surv
column of
dataspecifying a survival or time-to-event outcome, using theSurv()function. Right/left/interval censoring and left truncation (delayed entry) are supported.- trt_ref
reference treatment for the network, as a single integer, string, or factor. If not specified, a reasonable well-connected default will be chosen (see details).
- trt_class
column of
dataspecifying treatment classes, coded using integers, strings, or factors. By default, no classes are specified.
Value
An object of class nma_data
Details
By default, trt_ref = NULL and a network reference treatment will be chosen
that attempts to maximise computational efficiency and stability. If an
alternative reference treatment is chosen and the model runs slowly or has
low effective sample size (ESS) this may be the cause - try letting the
default reference treatment be used instead. Regardless of which treatment is
used as the network reference at the model fitting stage, results can be
transformed afterwards: see the trt_ref argument of
relative_effects() and predict.stan_nma().
All arguments specifying columns of data accept the following:
A column name as a character string, e.g.
study = "studyc"A bare column name, e.g.
study = studycdplyr::mutate()style semantics for inline variable transformations, e.g.study = paste(author, year)
See also
set_agd_arm() for arm-based aggregate data, set_agd_contrast()
for contrast-based aggregate data, and combine_network() for combining
several data sources in one network.
print.nma_data() for the print method displaying details of the
network, and plot.nma_data() for network plots.
Examples
# Set up network of plaque psoriasis IPD
head(plaque_psoriasis_ipd)
#> studyc trtc_long trtc trtn pasi75 pasi90 pasi100 age bmi pasi_w0
#> 1 IXORA-S Ixekizumab Q2W IXE_Q2W 2 1 1 1 62 38.6 15.8
#> 2 IXORA-S Ixekizumab Q2W IXE_Q2W 2 1 1 0 38 23.2 28.2
#> 3 IXORA-S Ixekizumab Q2W IXE_Q2W 2 1 1 0 54 27.5 13.2
#> 4 IXORA-S Ixekizumab Q2W IXE_Q2W 2 1 1 1 44 24.6 41.0
#> 5 IXORA-S Ixekizumab Q2W IXE_Q2W 2 1 1 0 44 28.3 15.2
#> 6 IXORA-S Ixekizumab Q2W IXE_Q2W 2 1 1 1 57 23.6 30.4
#> male bsa weight durnpso prevsys psa
#> 1 FALSE 13 111.2 8 TRUE TRUE
#> 2 FALSE 37 62.0 1 TRUE FALSE
#> 3 TRUE 13 83.5 38 TRUE FALSE
#> 4 FALSE 67 66.0 1 TRUE FALSE
#> 5 FALSE 10 92.7 23 TRUE FALSE
#> 6 FALSE 75 73.5 21 TRUE FALSE
pso_net <- set_ipd(plaque_psoriasis_ipd,
study = studyc,
trt = trtc,
r = pasi75)
# Print network details
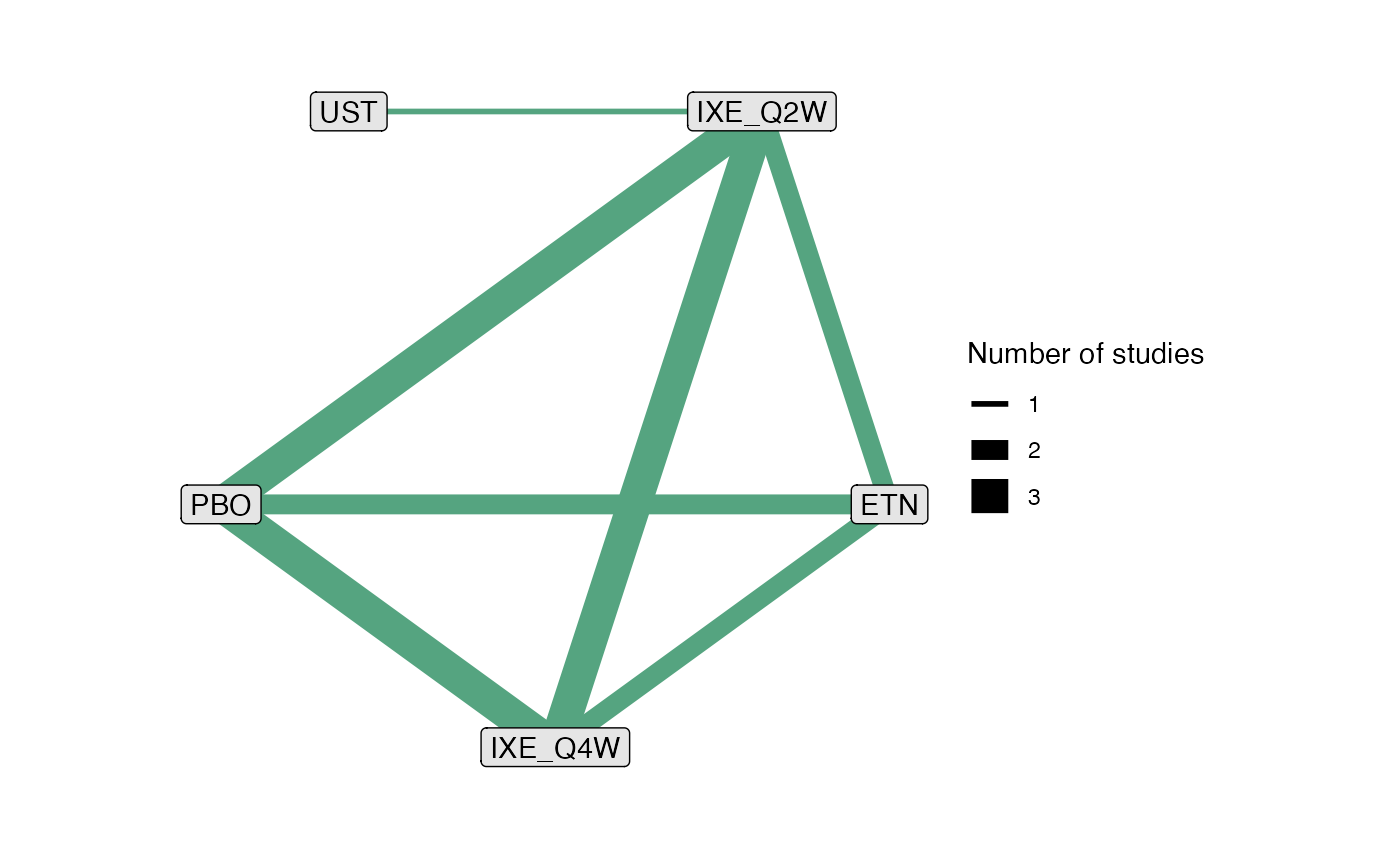
pso_net
#> A network with 4 IPD studies.
#>
#> ------------------------------------------------------------------- IPD studies ----
#> Study Treatment arms
#> IXORA-S 2: IXE_Q2W | UST
#> UNCOVER-1 3: IXE_Q2W | IXE_Q4W | PBO
#> UNCOVER-2 4: ETN | IXE_Q2W | IXE_Q4W | PBO
#> UNCOVER-3 4: ETN | IXE_Q2W | IXE_Q4W | PBO
#>
#> Outcome type: binary
#> ------------------------------------------------------------------------------------
#> Total number of treatments: 5
#> Total number of studies: 4
#> Reference treatment is: IXE_Q2W
#> Network is connected
# Plot network
plot(pso_net)
 # Setting a different reference treatment
set_ipd(plaque_psoriasis_ipd,
study = studyc,
trt = trtc,
r = pasi75,
trt_ref = "PBO")
#> A network with 4 IPD studies.
#>
#> ------------------------------------------------------------------- IPD studies ----
#> Study Treatment arms
#> IXORA-S 2: IXE_Q2W | UST
#> UNCOVER-1 3: IXE_Q2W | IXE_Q4W | PBO
#> UNCOVER-2 4: ETN | IXE_Q2W | IXE_Q4W | PBO
#> UNCOVER-3 4: ETN | IXE_Q2W | IXE_Q4W | PBO
#>
#> Outcome type: binary
#> ------------------------------------------------------------------------------------
#> Total number of treatments: 5
#> Total number of studies: 4
#> Reference treatment is: PBO
#> Network is connected
# Setting a different reference treatment
set_ipd(plaque_psoriasis_ipd,
study = studyc,
trt = trtc,
r = pasi75,
trt_ref = "PBO")
#> A network with 4 IPD studies.
#>
#> ------------------------------------------------------------------- IPD studies ----
#> Study Treatment arms
#> IXORA-S 2: IXE_Q2W | UST
#> UNCOVER-1 3: IXE_Q2W | IXE_Q4W | PBO
#> UNCOVER-2 4: ETN | IXE_Q2W | IXE_Q4W | PBO
#> UNCOVER-3 4: ETN | IXE_Q2W | IXE_Q4W | PBO
#>
#> Outcome type: binary
#> ------------------------------------------------------------------------------------
#> Total number of treatments: 5
#> Total number of studies: 4
#> Reference treatment is: PBO
#> Network is connected