Generate (population-average) relative treatment effects. If a ML-NMR or meta-regression model was fitted, these are specific to each study population.
Usage
relative_effects(
x,
newdata = NULL,
study = NULL,
all_contrasts = FALSE,
trt_ref = NULL,
probs = c(0.025, 0.25, 0.5, 0.75, 0.975),
predictive_distribution = FALSE,
summary = TRUE
)Arguments
- x
A
stan_nmaobject created bynma()- newdata
Only used if a regression model is fitted. A data frame of study details, one row per study, giving the covariate values at which to produce relative effects. Column names must match variables in the regression model. If
NULL, relative effects are produced for all studies in the network.- study
Column of
newdatawhich specifies study names, otherwise studies will be labelled by row number.- all_contrasts
Logical, generate estimates for all contrasts (
TRUE), or just the "basic" contrasts against the network reference treatment (FALSE)? DefaultFALSE.- trt_ref
Reference treatment to construct relative effects against, if
all_contrasts = FALSE. By default, relative effects will be against the network reference treatment. Coerced to character string.- probs
Numeric vector of quantiles of interest to present in computed summary, default
c(0.025, 0.25, 0.5, 0.75, 0.975)- predictive_distribution
Logical, when a random effects model has been fitted, should the predictive distribution for relative effects in a new study be returned? Default
FALSE.- summary
Logical, calculate posterior summaries? Default
TRUE.
Value
A nma_summary object if summary = TRUE, otherwise a list
containing a 3D MCMC array of samples and (for regression models) a data
frame of study information.
See also
plot.nma_summary() for plotting the relative effects.
Examples
## Smoking cessation
# \donttest{
# Run smoking RE NMA example if not already available
if (!exists("smk_fit_RE")) example("example_smk_re", run.donttest = TRUE)
# }
# \donttest{
# Produce relative effects
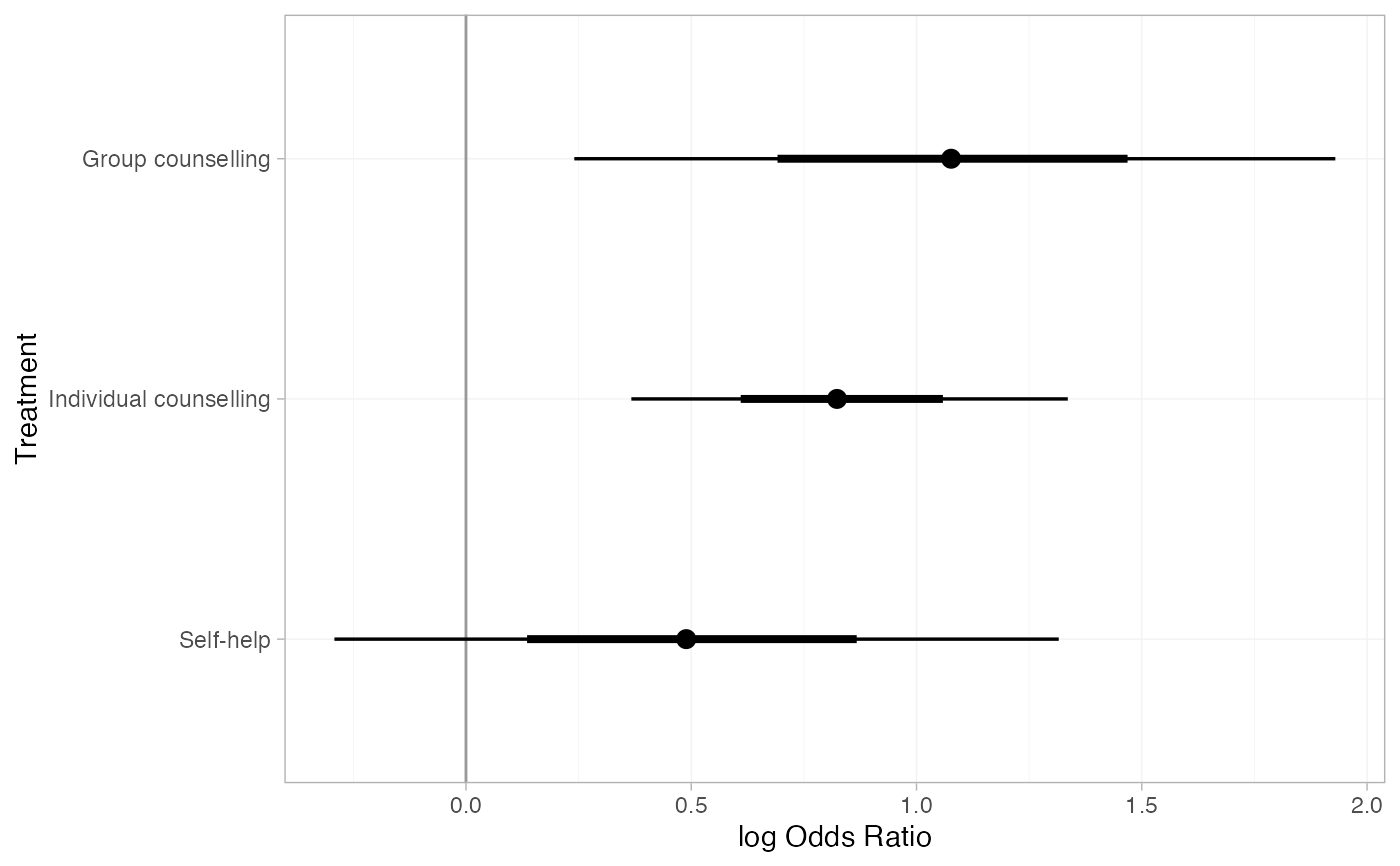
smk_releff_RE <- relative_effects(smk_fit_RE)
smk_releff_RE
#> mean sd 2.5% 25% 50% 75% 97.5% Bulk_ESS
#> d[Group counselling] 1.08 0.42 0.24 0.81 1.08 1.34 1.93 1927
#> d[Individual counselling] 0.83 0.24 0.37 0.67 0.82 0.99 1.34 1173
#> d[Self-help] 0.50 0.40 -0.29 0.24 0.49 0.76 1.32 1990
#> Tail_ESS Rhat
#> d[Group counselling] 2122 1
#> d[Individual counselling] 1735 1
#> d[Self-help] 2528 1
plot(smk_releff_RE, ref_line = 0)
 # Relative effects for all pairwise comparisons
relative_effects(smk_fit_RE, all_contrasts = TRUE)
#> mean sd 2.5% 25% 50%
#> d[Group counselling vs. No intervention] 1.08 0.42 0.24 0.81 1.08
#> d[Individual counselling vs. No intervention] 0.83 0.24 0.37 0.67 0.82
#> d[Self-help vs. No intervention] 0.50 0.40 -0.29 0.24 0.49
#> d[Individual counselling vs. Group counselling] -0.25 0.40 -1.03 -0.51 -0.25
#> d[Self-help vs. Group counselling] -0.58 0.47 -1.48 -0.90 -0.58
#> d[Self-help vs. Individual counselling] -0.33 0.40 -1.11 -0.58 -0.34
#> 75% 97.5% Bulk_ESS Tail_ESS
#> d[Group counselling vs. No intervention] 1.34 1.93 1927 2122
#> d[Individual counselling vs. No intervention] 0.99 1.34 1173 1735
#> d[Self-help vs. No intervention] 0.76 1.32 1990 2528
#> d[Individual counselling vs. Group counselling] 0.02 0.54 2698 2503
#> d[Self-help vs. Group counselling] -0.28 0.38 3019 2749
#> d[Self-help vs. Individual counselling] -0.07 0.47 2311 2468
#> Rhat
#> d[Group counselling vs. No intervention] 1
#> d[Individual counselling vs. No intervention] 1
#> d[Self-help vs. No intervention] 1
#> d[Individual counselling vs. Group counselling] 1
#> d[Self-help vs. Group counselling] 1
#> d[Self-help vs. Individual counselling] 1
# Relative effects against a different reference treatment
relative_effects(smk_fit_RE, trt_ref = "Self-help")
#> mean sd 2.5% 25% 50% 75% 97.5% Bulk_ESS
#> d[No intervention] -0.50 0.40 -1.32 -0.76 -0.49 -0.24 0.29 1990
#> d[Group counselling] 0.58 0.47 -0.38 0.28 0.58 0.90 1.48 3019
#> d[Individual counselling] 0.33 0.40 -0.47 0.07 0.34 0.58 1.11 2311
#> Tail_ESS Rhat
#> d[No intervention] 2528 1
#> d[Group counselling] 2749 1
#> d[Individual counselling] 2468 1
# Transforming to odds ratios
# We work with the array of relative effects samples
LOR_array <- as.array(smk_releff_RE)
OR_array <- exp(LOR_array)
# mcmc_array objects can be summarised to produce a nma_summary object
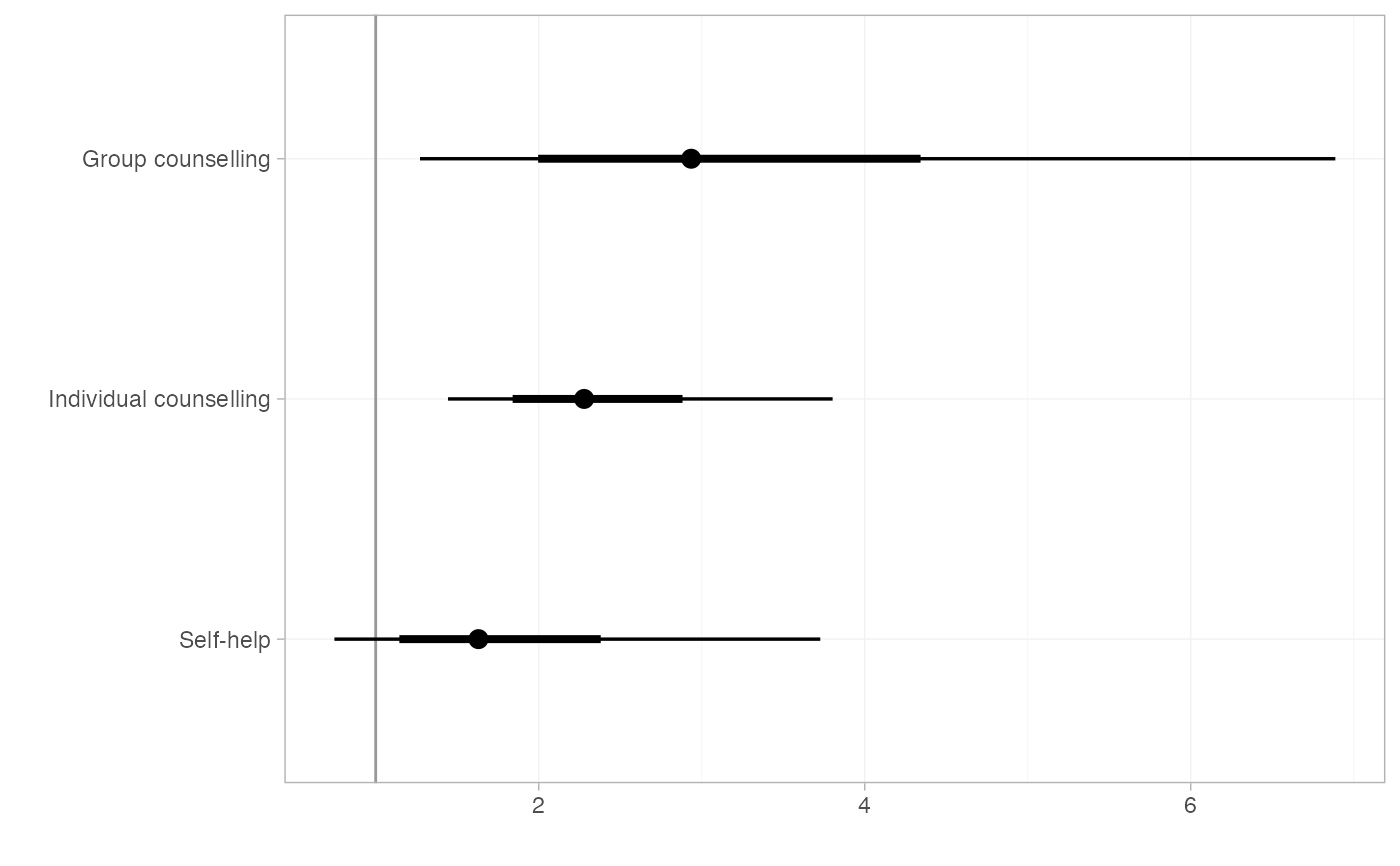
smk_OR_RE <- summary(OR_array)
# This can then be printed or plotted
smk_OR_RE
#> mean sd 2.5% 25% 50% 75% 97.5% Bulk_ESS Tail_ESS
#> d[Group counselling] 3.22 1.51 1.27 2.24 2.94 3.82 6.89 1927 2122
#> d[Individual counselling] 2.37 0.61 1.44 1.96 2.28 2.68 3.80 1173 1735
#> d[Self-help] 1.79 0.77 0.75 1.27 1.63 2.14 3.73 1990 2528
#> Rhat
#> d[Group counselling] 1
#> d[Individual counselling] 1
#> d[Self-help] 1
plot(smk_OR_RE, ref_line = 1)
# Relative effects for all pairwise comparisons
relative_effects(smk_fit_RE, all_contrasts = TRUE)
#> mean sd 2.5% 25% 50%
#> d[Group counselling vs. No intervention] 1.08 0.42 0.24 0.81 1.08
#> d[Individual counselling vs. No intervention] 0.83 0.24 0.37 0.67 0.82
#> d[Self-help vs. No intervention] 0.50 0.40 -0.29 0.24 0.49
#> d[Individual counselling vs. Group counselling] -0.25 0.40 -1.03 -0.51 -0.25
#> d[Self-help vs. Group counselling] -0.58 0.47 -1.48 -0.90 -0.58
#> d[Self-help vs. Individual counselling] -0.33 0.40 -1.11 -0.58 -0.34
#> 75% 97.5% Bulk_ESS Tail_ESS
#> d[Group counselling vs. No intervention] 1.34 1.93 1927 2122
#> d[Individual counselling vs. No intervention] 0.99 1.34 1173 1735
#> d[Self-help vs. No intervention] 0.76 1.32 1990 2528
#> d[Individual counselling vs. Group counselling] 0.02 0.54 2698 2503
#> d[Self-help vs. Group counselling] -0.28 0.38 3019 2749
#> d[Self-help vs. Individual counselling] -0.07 0.47 2311 2468
#> Rhat
#> d[Group counselling vs. No intervention] 1
#> d[Individual counselling vs. No intervention] 1
#> d[Self-help vs. No intervention] 1
#> d[Individual counselling vs. Group counselling] 1
#> d[Self-help vs. Group counselling] 1
#> d[Self-help vs. Individual counselling] 1
# Relative effects against a different reference treatment
relative_effects(smk_fit_RE, trt_ref = "Self-help")
#> mean sd 2.5% 25% 50% 75% 97.5% Bulk_ESS
#> d[No intervention] -0.50 0.40 -1.32 -0.76 -0.49 -0.24 0.29 1990
#> d[Group counselling] 0.58 0.47 -0.38 0.28 0.58 0.90 1.48 3019
#> d[Individual counselling] 0.33 0.40 -0.47 0.07 0.34 0.58 1.11 2311
#> Tail_ESS Rhat
#> d[No intervention] 2528 1
#> d[Group counselling] 2749 1
#> d[Individual counselling] 2468 1
# Transforming to odds ratios
# We work with the array of relative effects samples
LOR_array <- as.array(smk_releff_RE)
OR_array <- exp(LOR_array)
# mcmc_array objects can be summarised to produce a nma_summary object
smk_OR_RE <- summary(OR_array)
# This can then be printed or plotted
smk_OR_RE
#> mean sd 2.5% 25% 50% 75% 97.5% Bulk_ESS Tail_ESS
#> d[Group counselling] 3.22 1.51 1.27 2.24 2.94 3.82 6.89 1927 2122
#> d[Individual counselling] 2.37 0.61 1.44 1.96 2.28 2.68 3.80 1173 1735
#> d[Self-help] 1.79 0.77 0.75 1.27 1.63 2.14 3.73 1990 2528
#> Rhat
#> d[Group counselling] 1
#> d[Individual counselling] 1
#> d[Self-help] 1
plot(smk_OR_RE, ref_line = 1)
 # }
## Plaque psoriasis ML-NMR
# \donttest{
# Run plaque psoriasis ML-NMR example if not already available
if (!exists("pso_fit")) example("example_pso_mlnmr", run.donttest = TRUE)
# }
# \donttest{
# Produce population-adjusted relative effects for all study populations in
# the network
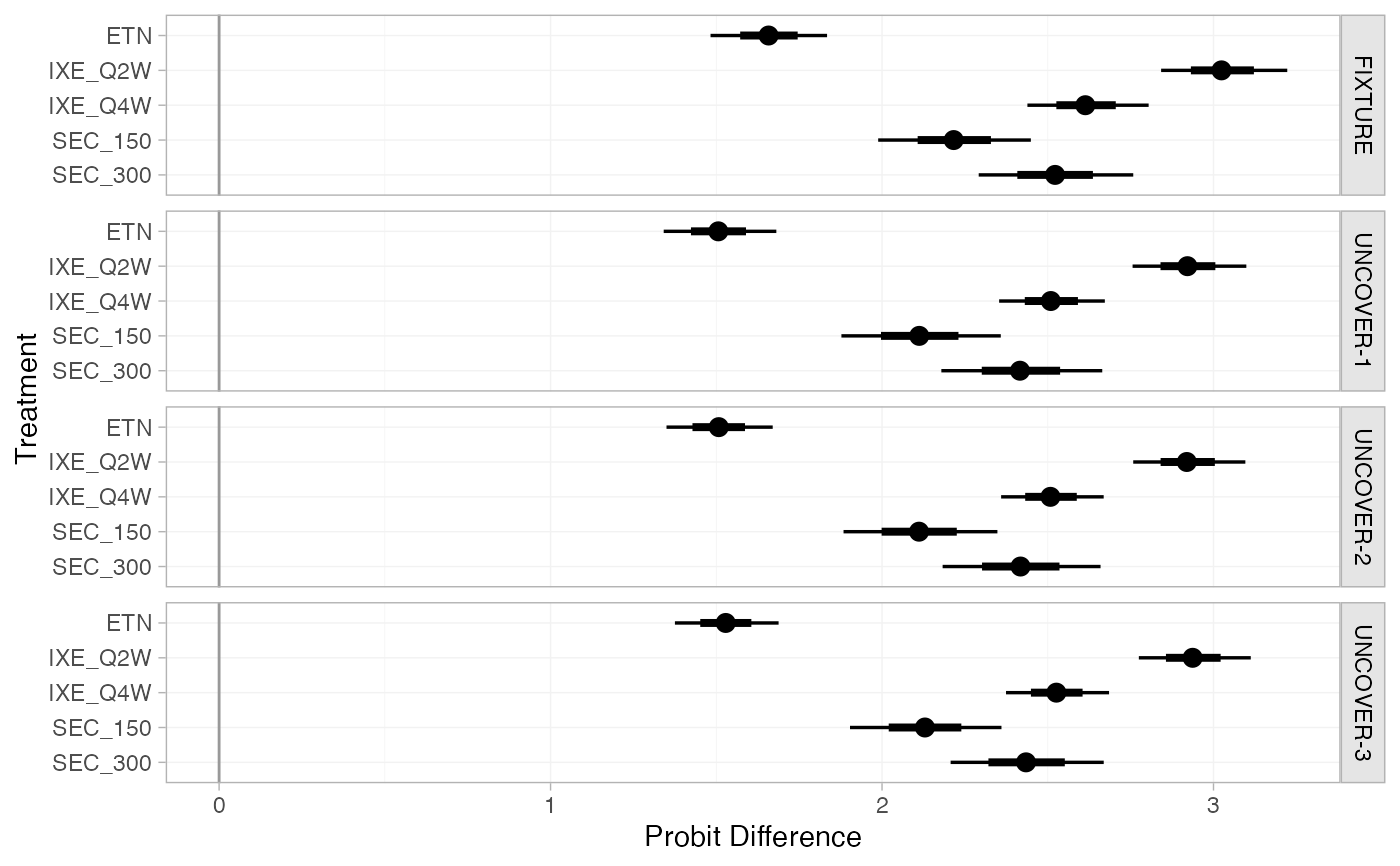
pso_releff <- relative_effects(pso_fit)
pso_releff
#> ---------------------------------------------------------------- Study: FIXTURE ----
#>
#> Covariate values:
#> durnpso prevsys bsa weight psa
#> 1.6 0.62 0.34 8.34 0.14
#>
#> mean sd 2.5% 25% 50% 75% 97.5% Bulk_ESS Tail_ESS Rhat
#> d[FIXTURE: ETN] 1.66 0.09 1.48 1.60 1.66 1.72 1.83 4513 3345 1
#> d[FIXTURE: IXE_Q2W] 3.03 0.10 2.84 2.96 3.02 3.09 3.22 5525 3008 1
#> d[FIXTURE: IXE_Q4W] 2.61 0.09 2.44 2.55 2.61 2.68 2.80 5577 3082 1
#> d[FIXTURE: SEC_150] 2.22 0.12 1.99 2.14 2.22 2.29 2.45 4577 3514 1
#> d[FIXTURE: SEC_300] 2.52 0.12 2.29 2.44 2.52 2.60 2.76 5349 3031 1
#>
#> -------------------------------------------------------------- Study: UNCOVER-1 ----
#>
#> Covariate values:
#> durnpso prevsys bsa weight psa
#> 2 0.73 0.28 9.24 0.28
#>
#> mean sd 2.5% 25% 50% 75% 97.5% Bulk_ESS Tail_ESS
#> d[UNCOVER-1: ETN] 1.51 0.09 1.34 1.45 1.51 1.56 1.68 3949 3556
#> d[UNCOVER-1: IXE_Q2W] 2.92 0.09 2.76 2.86 2.92 2.98 3.10 5578 3398
#> d[UNCOVER-1: IXE_Q4W] 2.51 0.08 2.35 2.45 2.51 2.57 2.67 5584 3028
#> d[UNCOVER-1: SEC_150] 2.11 0.12 1.88 2.03 2.11 2.20 2.36 4648 3485
#> d[UNCOVER-1: SEC_300] 2.42 0.12 2.18 2.33 2.42 2.50 2.66 5052 3333
#> Rhat
#> d[UNCOVER-1: ETN] 1
#> d[UNCOVER-1: IXE_Q2W] 1
#> d[UNCOVER-1: IXE_Q4W] 1
#> d[UNCOVER-1: SEC_150] 1
#> d[UNCOVER-1: SEC_300] 1
#>
#> -------------------------------------------------------------- Study: UNCOVER-2 ----
#>
#> Covariate values:
#> durnpso prevsys bsa weight psa
#> 1.87 0.64 0.27 9.17 0.24
#>
#> mean sd 2.5% 25% 50% 75% 97.5% Bulk_ESS Tail_ESS
#> d[UNCOVER-2: ETN] 1.51 0.08 1.35 1.45 1.51 1.56 1.67 3931 3260
#> d[UNCOVER-2: IXE_Q2W] 2.92 0.09 2.76 2.86 2.92 2.98 3.10 5489 3424
#> d[UNCOVER-2: IXE_Q4W] 2.51 0.08 2.36 2.45 2.51 2.56 2.67 5465 3262
#> d[UNCOVER-2: SEC_150] 2.11 0.12 1.88 2.03 2.11 2.19 2.35 4745 3537
#> d[UNCOVER-2: SEC_300] 2.42 0.12 2.18 2.34 2.42 2.50 2.66 5133 3189
#> Rhat
#> d[UNCOVER-2: ETN] 1
#> d[UNCOVER-2: IXE_Q2W] 1
#> d[UNCOVER-2: IXE_Q4W] 1
#> d[UNCOVER-2: SEC_150] 1
#> d[UNCOVER-2: SEC_300] 1
#>
#> -------------------------------------------------------------- Study: UNCOVER-3 ----
#>
#> Covariate values:
#> durnpso prevsys bsa weight psa
#> 1.78 0.59 0.28 9.01 0.2
#>
#> mean sd 2.5% 25% 50% 75% 97.5% Bulk_ESS Tail_ESS
#> d[UNCOVER-3: ETN] 1.53 0.08 1.38 1.47 1.53 1.58 1.69 3948 3433
#> d[UNCOVER-3: IXE_Q2W] 2.94 0.09 2.77 2.88 2.94 3.00 3.11 5582 3334
#> d[UNCOVER-3: IXE_Q4W] 2.53 0.08 2.37 2.47 2.53 2.58 2.68 5325 3410
#> d[UNCOVER-3: SEC_150] 2.13 0.12 1.90 2.05 2.13 2.21 2.36 4752 3428
#> d[UNCOVER-3: SEC_300] 2.43 0.12 2.21 2.35 2.43 2.51 2.67 5092 3372
#> Rhat
#> d[UNCOVER-3: ETN] 1
#> d[UNCOVER-3: IXE_Q2W] 1
#> d[UNCOVER-3: IXE_Q4W] 1
#> d[UNCOVER-3: SEC_150] 1
#> d[UNCOVER-3: SEC_300] 1
#>
plot(pso_releff, ref_line = 0)
# }
## Plaque psoriasis ML-NMR
# \donttest{
# Run plaque psoriasis ML-NMR example if not already available
if (!exists("pso_fit")) example("example_pso_mlnmr", run.donttest = TRUE)
# }
# \donttest{
# Produce population-adjusted relative effects for all study populations in
# the network
pso_releff <- relative_effects(pso_fit)
pso_releff
#> ---------------------------------------------------------------- Study: FIXTURE ----
#>
#> Covariate values:
#> durnpso prevsys bsa weight psa
#> 1.6 0.62 0.34 8.34 0.14
#>
#> mean sd 2.5% 25% 50% 75% 97.5% Bulk_ESS Tail_ESS Rhat
#> d[FIXTURE: ETN] 1.66 0.09 1.48 1.60 1.66 1.72 1.83 4513 3345 1
#> d[FIXTURE: IXE_Q2W] 3.03 0.10 2.84 2.96 3.02 3.09 3.22 5525 3008 1
#> d[FIXTURE: IXE_Q4W] 2.61 0.09 2.44 2.55 2.61 2.68 2.80 5577 3082 1
#> d[FIXTURE: SEC_150] 2.22 0.12 1.99 2.14 2.22 2.29 2.45 4577 3514 1
#> d[FIXTURE: SEC_300] 2.52 0.12 2.29 2.44 2.52 2.60 2.76 5349 3031 1
#>
#> -------------------------------------------------------------- Study: UNCOVER-1 ----
#>
#> Covariate values:
#> durnpso prevsys bsa weight psa
#> 2 0.73 0.28 9.24 0.28
#>
#> mean sd 2.5% 25% 50% 75% 97.5% Bulk_ESS Tail_ESS
#> d[UNCOVER-1: ETN] 1.51 0.09 1.34 1.45 1.51 1.56 1.68 3949 3556
#> d[UNCOVER-1: IXE_Q2W] 2.92 0.09 2.76 2.86 2.92 2.98 3.10 5578 3398
#> d[UNCOVER-1: IXE_Q4W] 2.51 0.08 2.35 2.45 2.51 2.57 2.67 5584 3028
#> d[UNCOVER-1: SEC_150] 2.11 0.12 1.88 2.03 2.11 2.20 2.36 4648 3485
#> d[UNCOVER-1: SEC_300] 2.42 0.12 2.18 2.33 2.42 2.50 2.66 5052 3333
#> Rhat
#> d[UNCOVER-1: ETN] 1
#> d[UNCOVER-1: IXE_Q2W] 1
#> d[UNCOVER-1: IXE_Q4W] 1
#> d[UNCOVER-1: SEC_150] 1
#> d[UNCOVER-1: SEC_300] 1
#>
#> -------------------------------------------------------------- Study: UNCOVER-2 ----
#>
#> Covariate values:
#> durnpso prevsys bsa weight psa
#> 1.87 0.64 0.27 9.17 0.24
#>
#> mean sd 2.5% 25% 50% 75% 97.5% Bulk_ESS Tail_ESS
#> d[UNCOVER-2: ETN] 1.51 0.08 1.35 1.45 1.51 1.56 1.67 3931 3260
#> d[UNCOVER-2: IXE_Q2W] 2.92 0.09 2.76 2.86 2.92 2.98 3.10 5489 3424
#> d[UNCOVER-2: IXE_Q4W] 2.51 0.08 2.36 2.45 2.51 2.56 2.67 5465 3262
#> d[UNCOVER-2: SEC_150] 2.11 0.12 1.88 2.03 2.11 2.19 2.35 4745 3537
#> d[UNCOVER-2: SEC_300] 2.42 0.12 2.18 2.34 2.42 2.50 2.66 5133 3189
#> Rhat
#> d[UNCOVER-2: ETN] 1
#> d[UNCOVER-2: IXE_Q2W] 1
#> d[UNCOVER-2: IXE_Q4W] 1
#> d[UNCOVER-2: SEC_150] 1
#> d[UNCOVER-2: SEC_300] 1
#>
#> -------------------------------------------------------------- Study: UNCOVER-3 ----
#>
#> Covariate values:
#> durnpso prevsys bsa weight psa
#> 1.78 0.59 0.28 9.01 0.2
#>
#> mean sd 2.5% 25% 50% 75% 97.5% Bulk_ESS Tail_ESS
#> d[UNCOVER-3: ETN] 1.53 0.08 1.38 1.47 1.53 1.58 1.69 3948 3433
#> d[UNCOVER-3: IXE_Q2W] 2.94 0.09 2.77 2.88 2.94 3.00 3.11 5582 3334
#> d[UNCOVER-3: IXE_Q4W] 2.53 0.08 2.37 2.47 2.53 2.58 2.68 5325 3410
#> d[UNCOVER-3: SEC_150] 2.13 0.12 1.90 2.05 2.13 2.21 2.36 4752 3428
#> d[UNCOVER-3: SEC_300] 2.43 0.12 2.21 2.35 2.43 2.51 2.67 5092 3372
#> Rhat
#> d[UNCOVER-3: ETN] 1
#> d[UNCOVER-3: IXE_Q2W] 1
#> d[UNCOVER-3: IXE_Q4W] 1
#> d[UNCOVER-3: SEC_150] 1
#> d[UNCOVER-3: SEC_300] 1
#>
plot(pso_releff, ref_line = 0)
 # Produce population-adjusted relative effects for a different target
# population
new_agd_means <- data.frame(
bsa = 0.6,
prevsys = 0.1,
psa = 0.2,
weight = 10,
durnpso = 3)
relative_effects(pso_fit, newdata = new_agd_means)
#> ------------------------------------------------------------------ Study: New 1 ----
#>
#> Covariate values:
#> durnpso prevsys bsa weight psa
#> 3 0.1 0.6 10 0.2
#>
#> mean sd 2.5% 25% 50% 75% 97.5% Bulk_ESS Tail_ESS Rhat
#> d[New 1: ETN] 1.25 0.23 0.80 1.10 1.25 1.40 1.71 6553 3024 1
#> d[New 1: IXE_Q2W] 2.88 0.23 2.46 2.73 2.88 3.04 3.34 6972 3082 1
#> d[New 1: IXE_Q4W] 2.47 0.22 2.05 2.32 2.47 2.62 2.91 6962 3203 1
#> d[New 1: SEC_150] 2.08 0.23 1.63 1.92 2.07 2.23 2.53 6516 2947 1
#> d[New 1: SEC_300] 2.38 0.23 1.95 2.22 2.38 2.53 2.83 7079 3406 1
#>
# }
# Produce population-adjusted relative effects for a different target
# population
new_agd_means <- data.frame(
bsa = 0.6,
prevsys = 0.1,
psa = 0.2,
weight = 10,
durnpso = 3)
relative_effects(pso_fit, newdata = new_agd_means)
#> ------------------------------------------------------------------ Study: New 1 ----
#>
#> Covariate values:
#> durnpso prevsys bsa weight psa
#> 3 0.1 0.6 10 0.2
#>
#> mean sd 2.5% 25% 50% 75% 97.5% Bulk_ESS Tail_ESS Rhat
#> d[New 1: ETN] 1.25 0.23 0.80 1.10 1.25 1.40 1.71 6553 3024 1
#> d[New 1: IXE_Q2W] 2.88 0.23 2.46 2.73 2.88 3.04 3.34 6972 3082 1
#> d[New 1: IXE_Q4W] 2.47 0.22 2.05 2.32 2.47 2.62 2.91 6962 3203 1
#> d[New 1: SEC_150] 2.08 0.23 1.63 1.92 2.07 2.23 2.53 6516 2947 1
#> d[New 1: SEC_300] 2.38 0.23 1.95 2.22 2.38 2.53 2.83 7079 3406 1
#>
# }