Set up a network containing arm-based aggregate data (AgD), such as event
counts or mean outcomes on each arm. Multiple data sources may be combined
once created using combine_network().
Usage
set_agd_arm(
data,
study,
trt,
y = NULL,
se = NULL,
r = NULL,
n = NULL,
E = NULL,
sample_size = NULL,
trt_ref = NULL,
trt_class = NULL
)Arguments
- data
a data frame
- study
column of
dataspecifying the studies, coded using integers, strings, or factors- trt
column of
dataspecifying treatments, coded using integers, strings, or factors- y
column of
dataspecifying a continuous outcome- se
column of
dataspecifying the standard error for a continuous outcome- r
column of
dataspecifying a binary or Binomial outcome count- n
column of
dataspecifying Binomial outcome numerator- E
column of
dataspecifying the total time at risk for Poisson outcomes- sample_size
column of
datagiving the sample size in each arm. Optional, see details.- trt_ref
reference treatment for the network, as a single integer, string, or factor. If not specified, a reasonable well-connected default will be chosen (see details).
- trt_class
column of
dataspecifying treatment classes, coded using integers, strings, or factors. By default, no classes are specified.
Value
An object of class nma_data
Details
By default, trt_ref = NULL and a network reference treatment will be chosen
that attempts to maximise computational efficiency and stability. If an
alternative reference treatment is chosen and the model runs slowly or has
low effective sample size (ESS) this may be the cause - try letting the
default reference treatment be used instead. Regardless of which treatment is
used as the network reference at the model fitting stage, results can be
transformed afterwards: see the trt_ref argument of
relative_effects() and predict.stan_nma().
The sample_size argument is optional, but when specified:
Enables automatic centering of predictors (
center = TRUE) innma()when a regression model is given for a network combining IPD and AgDEnables production of study-specific relative effects, rank probabilities, etc. for studies in the network when a regression model is given
Nodes in
plot.nma_data()may be weighted by sample size
If a Binomial outcome is specified and sample_size is omitted, n will be
used as the sample size by default. If a Multinomial outcome is specified and
sample_size is omitted, the sample size will be determined automatically
from the supplied counts by default.
All arguments specifying columns of data accept the following:
A column name as a character string, e.g.
study = "studyc"A bare column name, e.g.
study = studycdplyr::mutate()style semantics for inline variable transformations, e.g.study = paste(author, year)
See also
set_ipd() for individual patient data, set_agd_contrast() for
contrast-based aggregate data, and combine_network() for combining
several data sources in one network.
print.nma_data() for the print method displaying details of the
network, and plot.nma_data() for network plots.
Examples
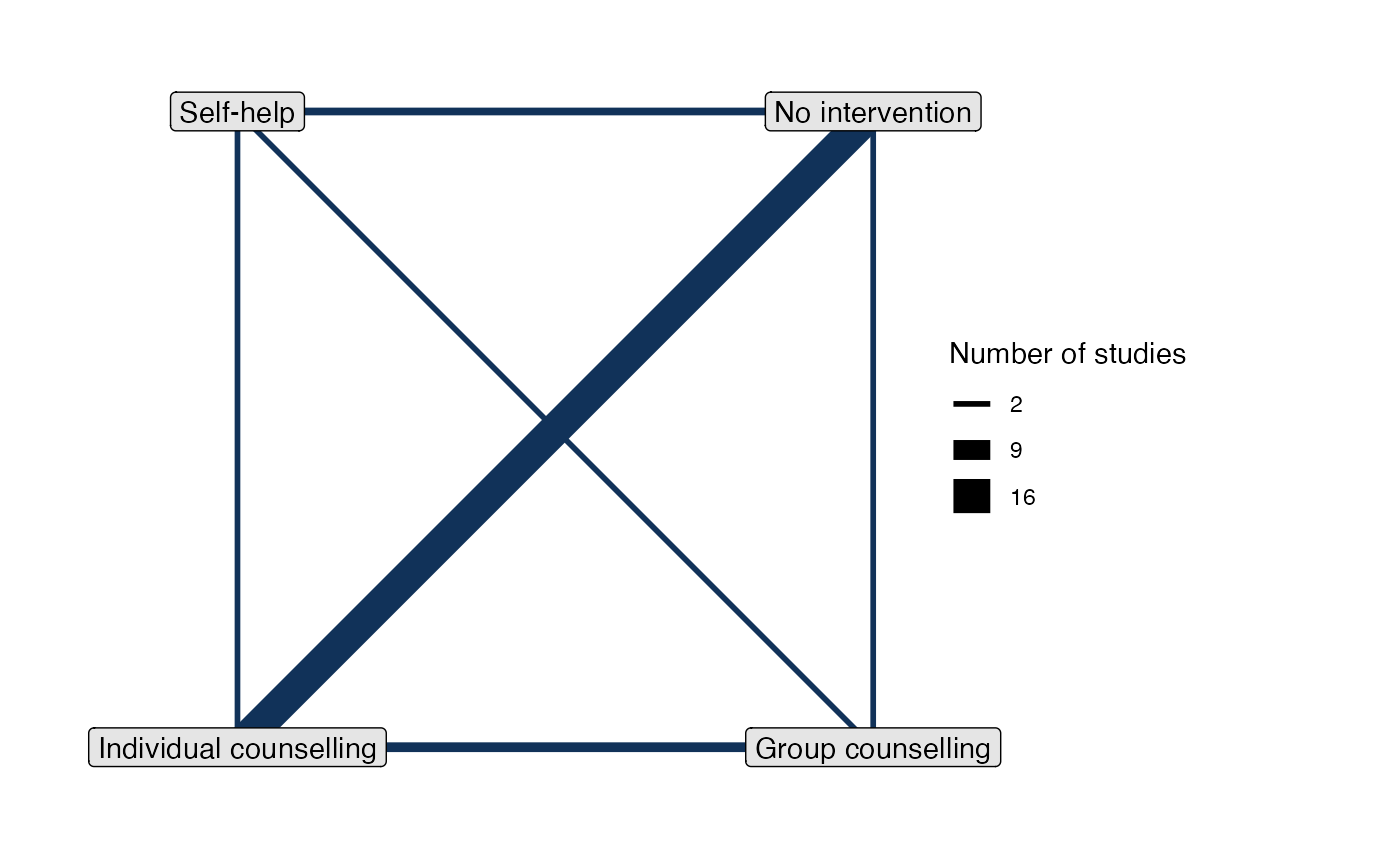
# Set up network of smoking cessation data
head(smoking)
#> studyn trtn trtc r n
#> 1 1 1 No intervention 9 140
#> 2 1 3 Individual counselling 23 140
#> 3 1 4 Group counselling 10 138
#> 4 2 2 Self-help 11 78
#> 5 2 3 Individual counselling 12 85
#> 6 2 4 Group counselling 29 170
smk_net <- set_agd_arm(smoking,
study = studyn,
trt = trtc,
r = r,
n = n,
trt_ref = "No intervention")
# Print details
smk_net
#> A network with 24 AgD studies (arm-based).
#>
#> ------------------------------------------------------- AgD studies (arm-based) ----
#> Study Treatment arms
#> 1 3: No intervention | Group counselling | Individual counselling
#> 2 3: Group counselling | Individual counselling | Self-help
#> 3 2: No intervention | Individual counselling
#> 4 2: No intervention | Individual counselling
#> 5 2: No intervention | Individual counselling
#> 6 2: No intervention | Individual counselling
#> 7 2: No intervention | Individual counselling
#> 8 2: No intervention | Individual counselling
#> 9 2: No intervention | Individual counselling
#> 10 2: No intervention | Self-help
#> ... plus 14 more studies
#>
#> Outcome type: count
#> ------------------------------------------------------------------------------------
#> Total number of treatments: 4
#> Total number of studies: 24
#> Reference treatment is: No intervention
#> Network is connected
# Plot network
plot(smk_net)