3D MCMC arrays (Iterations, Chains, Parameters) are produced by as.array()
methods applied to stan_nma or nma_summary objects.
Value
The summary() method returns a nma_summary object, the print()
method returns x invisibly. The names() method returns a character
vector of parameter names, and names()<- returns the object with updated
parameter names. The plot() method is a shortcut for
plot(summary(x), ...), passing all arguments on to plot.nma_summary().
Examples
## Smoking cessation
# \donttest{
# Run smoking RE NMA example if not already available
if (!exists("smk_fit_RE")) example("example_smk_re", run.donttest = TRUE)
# }
# \donttest{
# Working with arrays of posterior draws (as mcmc_array objects) is
# convenient when transforming parameters
# Transforming log odds ratios to odds ratios
LOR_array <- as.array(relative_effects(smk_fit_RE))
OR_array <- exp(LOR_array)
# mcmc_array objects can be summarised to produce a nma_summary object
smk_OR_RE <- summary(OR_array)
# This can then be printed or plotted
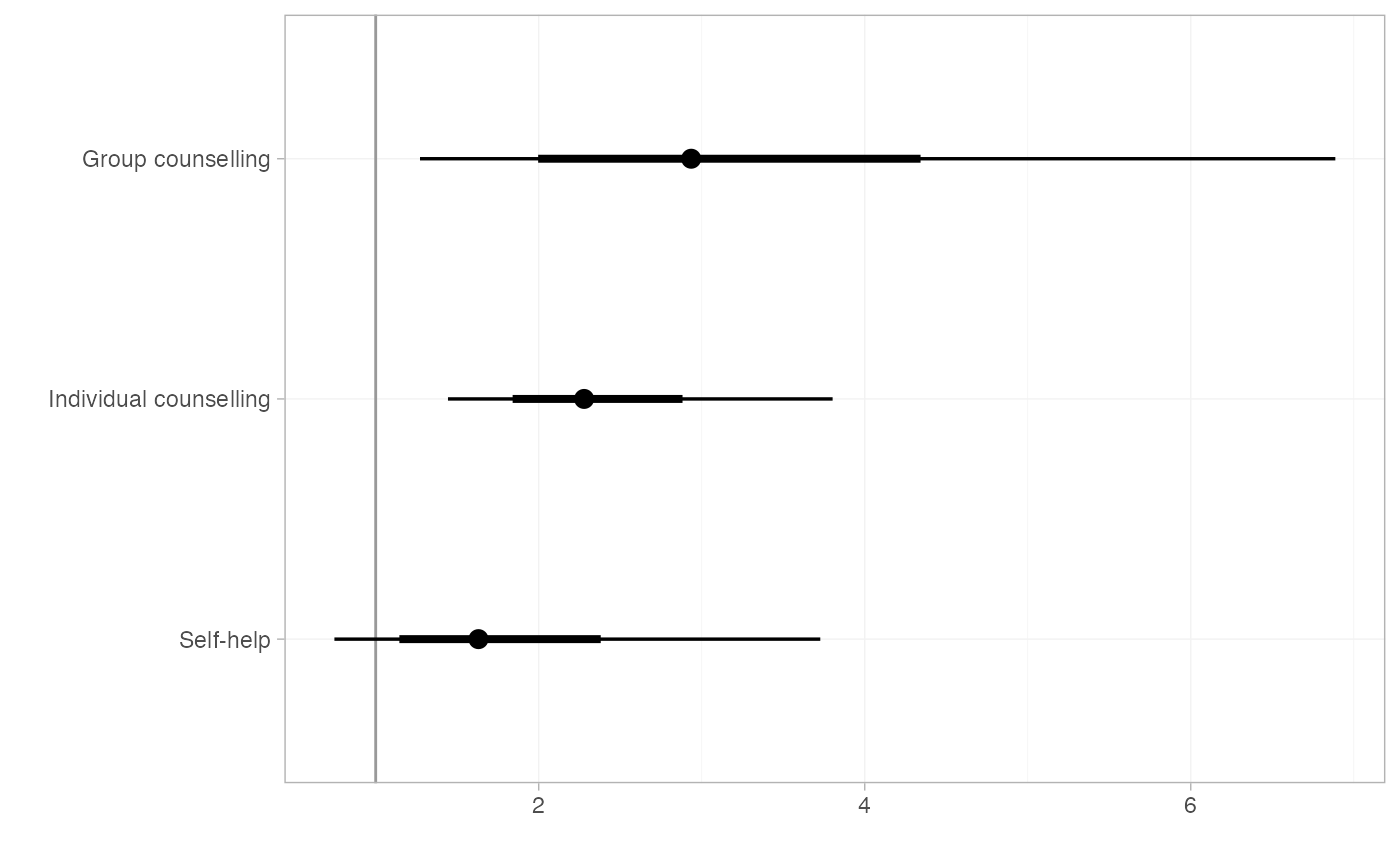
smk_OR_RE
#> mean sd 2.5% 25% 50% 75% 97.5% Bulk_ESS Tail_ESS
#> d[Group counselling] 3.22 1.51 1.27 2.24 2.94 3.82 6.89 1927 2122
#> d[Individual counselling] 2.37 0.61 1.44 1.96 2.28 2.68 3.80 1173 1735
#> d[Self-help] 1.79 0.77 0.75 1.27 1.63 2.14 3.73 1990 2528
#> Rhat
#> d[Group counselling] 1
#> d[Individual counselling] 1
#> d[Self-help] 1
plot(smk_OR_RE, ref_line = 1)
 # Transforming heterogeneity SD to variance
tau_array <- as.array(smk_fit_RE, pars = "tau")
tausq_array <- tau_array^2
# Correct parameter names
names(tausq_array) <- "tausq"
# Summarise
summary(tausq_array)
#> mean sd 2.5% 25% 50% 75% 97.5% Bulk_ESS Tail_ESS Rhat
#> tausq 0.71 0.32 0.29 0.49 0.64 0.87 1.53 1325 2047 1
# }
# Transforming heterogeneity SD to variance
tau_array <- as.array(smk_fit_RE, pars = "tau")
tausq_array <- tau_array^2
# Correct parameter names
names(tausq_array) <- "tausq"
# Summarise
summary(tausq_array)
#> mean sd 2.5% 25% 50% 75% 97.5% Bulk_ESS Tail_ESS Rhat
#> tausq 0.71 0.32 0.29 0.49 0.64 0.87 1.53 1325 2047 1
# }