Calculate the DIC for a model fitted using the nma() function.
Usage
dic(x, penalty = c("pD", "pV"), ...)Arguments
- x
A fitted model object, inheriting class stan_nma
- penalty
The method for estimating the effective number of parameters, used to penalise model fit in the DIC. Either
"pD"(the default), or"pV". For survival likelihoods only"pV"is currently available.- ...
Other arguments (not used)
Value
A nma_dic object.
See also
print.nma_dic() for printing details, plot.nma_dic() for
producing plots of residual deviance contributions.
Examples
## Smoking cessation
# \donttest{
# Run smoking FE NMA example if not already available
if (!exists("smk_fit_FE")) example("example_smk_fe", run.donttest = TRUE)
# }
# \donttest{
# Run smoking RE NMA example if not already available
if (!exists("smk_fit_RE")) example("example_smk_re", run.donttest = TRUE)
# }
# \donttest{
# Compare DIC of FE and RE models
(smk_dic_FE <- dic(smk_fit_FE))
#> Residual deviance: 267.1 (on 50 data points)
#> pD: 26.9
#> DIC: 293.9
(smk_dic_RE <- dic(smk_fit_RE)) # substantially better fit
#> Residual deviance: 54.5 (on 50 data points)
#> pD: 44
#> DIC: 98.4
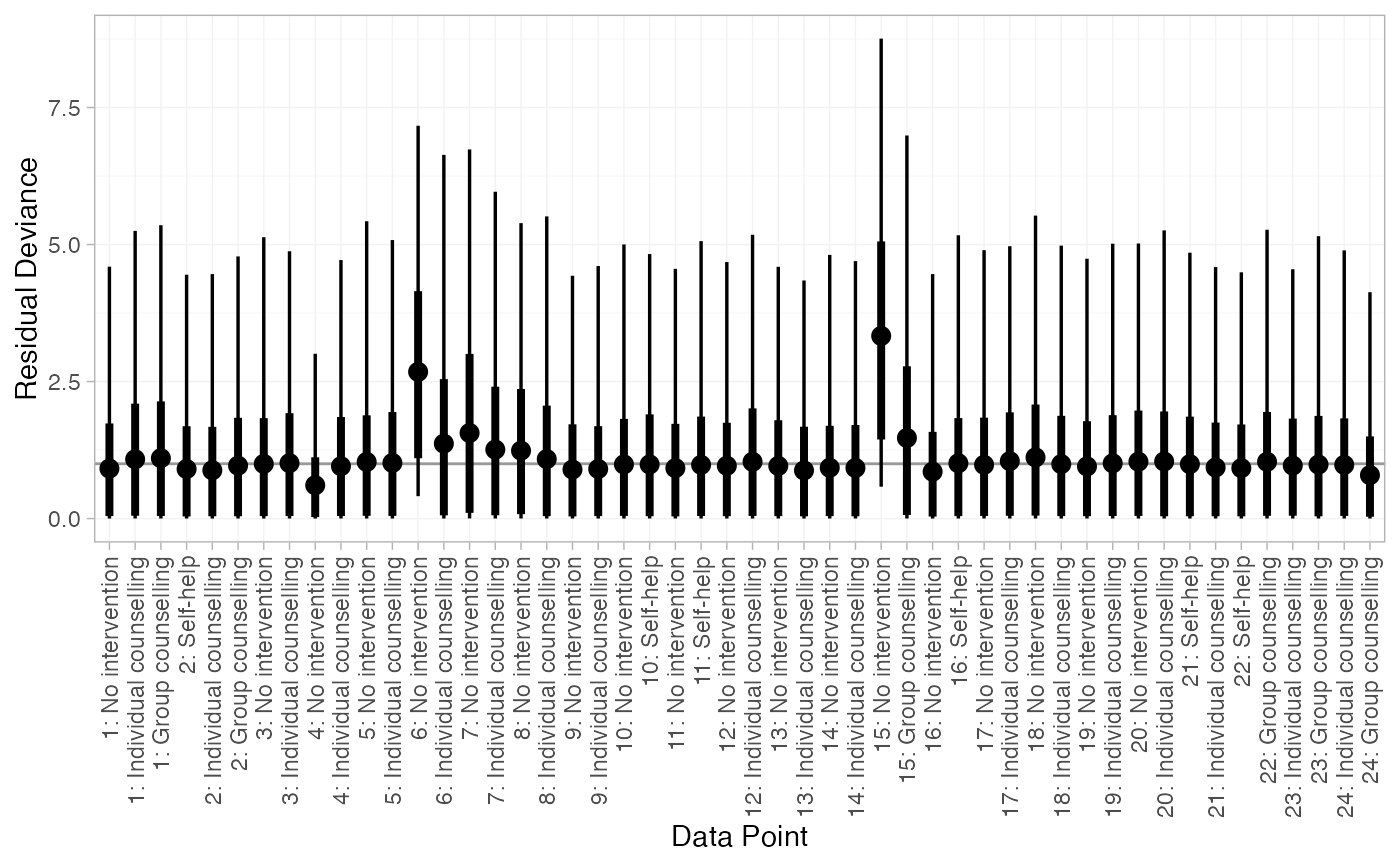
# Plot residual deviance contributions under RE model
plot(smk_dic_RE)
 # Check for inconsistency using UME model
# }
# \donttest{
# Run smoking UME NMA example if not already available
if (!exists("smk_fit_RE_UME")) example("example_smk_ume", run.donttest = TRUE)
# }
# \donttest{
# Compare DIC
smk_dic_RE
#> Residual deviance: 54.5 (on 50 data points)
#> pD: 44
#> DIC: 98.4
(smk_dic_RE_UME <- dic(smk_fit_RE_UME)) # no difference in fit
#> Residual deviance: 54 (on 50 data points)
#> pD: 45.4
#> DIC: 99.5
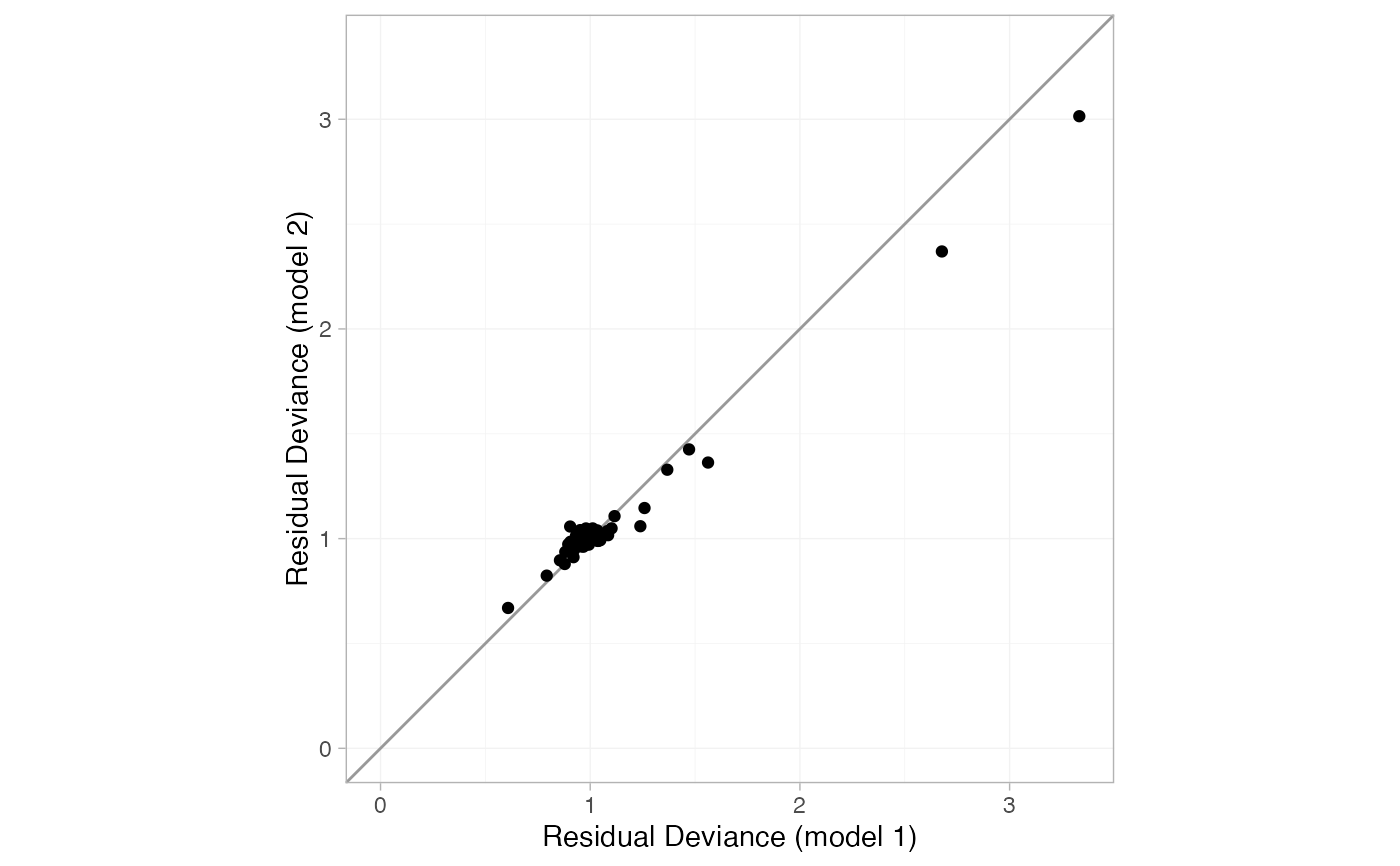
# Compare residual deviance contributions
plot(smk_dic_RE, smk_dic_RE_UME, show_uncertainty = FALSE)
# Check for inconsistency using UME model
# }
# \donttest{
# Run smoking UME NMA example if not already available
if (!exists("smk_fit_RE_UME")) example("example_smk_ume", run.donttest = TRUE)
# }
# \donttest{
# Compare DIC
smk_dic_RE
#> Residual deviance: 54.5 (on 50 data points)
#> pD: 44
#> DIC: 98.4
(smk_dic_RE_UME <- dic(smk_fit_RE_UME)) # no difference in fit
#> Residual deviance: 54 (on 50 data points)
#> pD: 45.4
#> DIC: 99.5
# Compare residual deviance contributions
plot(smk_dic_RE, smk_dic_RE_UME, show_uncertainty = FALSE)
 # }
# }